nf-core/genomeassembler
Assembly and scaffolding of haploid / unphased genomes from long ONT or PacBio HiFi reads
Introduction
nf-core/genomeassembler is a bioinformatics pipeline that carries out genome assembly, polishing and scaffolding from long reads (ONT or pacbio). Assembly can be done via flye or hifiasm, polishing can be carried out with medaka (ONT), or pilon (requires short-reads), and scaffolding can be done using LINKS, Longstitch, or RagTag (if a reference is available). Quality control includes BUSCO, QUAST and merqury (requires short-reads).
Currently, this pipeline does not implement phasing of polyploid genomes or HiC scaffolding.
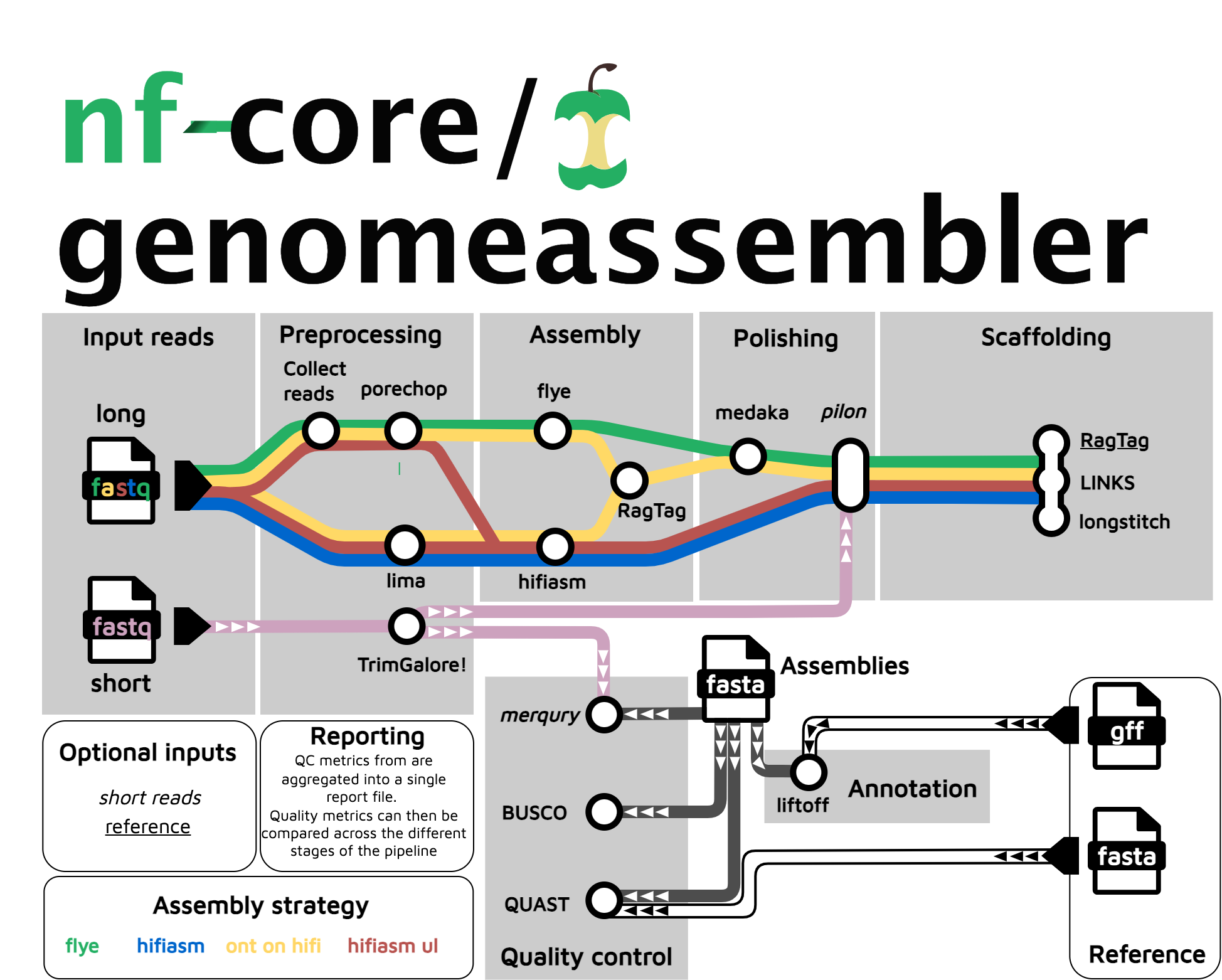
Usage
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
First, prepare a samplesheet with your input data that looks as follows:
samplesheet.csv:
sample,ontreads,hifireads,ref_fasta,ref_gff,shortread_F,shortread_R,paired
sampleName,ontreads.fa.gz,hifireads.fa.gz,assembly.fasta.gz,reference.fasta,reference.gff,short_F1.fastq,short_F2.fastq,trueEach row represents one genome to be assembled. sample should contain the name of the sample, ontreads should contain a path to ONT reads (fastq.gz), hifireads a path to HiFi reads (fastq.gz), ref_fasta and ref_gff contain reference genome fasta and annotations. shortread_F and shortread_R contain paths to short-read data, paired indicates if short-reads are paired. Columns can be omitted if they contain no data, with the exception of shortread_R, which needs to be present if shortread_F is there, even if it is empty.
Now, you can run the pipeline using:
nextflow run nf-core/genomeassembler \
-profile <docker/singularity/.../institute> \
--input samplesheet.csv \
--outdir <OUTDIR>Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those provided by the -c Nextflow option can be used to provide any configuration except for parameters; see docs.
For more details and further functionality, please refer to the usage documentation and the parameter documentation.
Pipeline output
To see the results of an example test run with a full size dataset refer to the results tab on the nf-core website pipeline page. For more details about the output files and reports, please refer to the output documentation.
Credits
nf-core/genomeassembler was originally written by Niklas Schandry, of the Faculty of Biology of the Ludwig-Maximilians University (LMU) in Munich, Germany.
I thank the following people for their extensive assistance and constructive reviews during the development of this pipeline:
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #genomeassembler channel (you can join with this invite).
Citations
If you use nf-core/genomeassembler for your analysis, please cite it using the following doi: 10.5281/zenodo.14986998
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.